Number of entries in biological sequence databases
What you should know about this indicator
- Biological sequence data includes data such as DNA and RNA sequences, amino acid sequences of proteins, and three-dimensional structures of proteins and other molecules found in living organisms. These organisms can be bacteria, viruses, plants, animals and humans.
- Researchers use this data to better understand the biology of organisms, including the functions of genes and proteins and how they interact. This knowledge can then be applied to e.g. develop new drugs and treatments for illnesses or use biotechnology to improve agriculture and environmental science.
- This dataset provides an overview of the growth of key biological sequence databases over time.
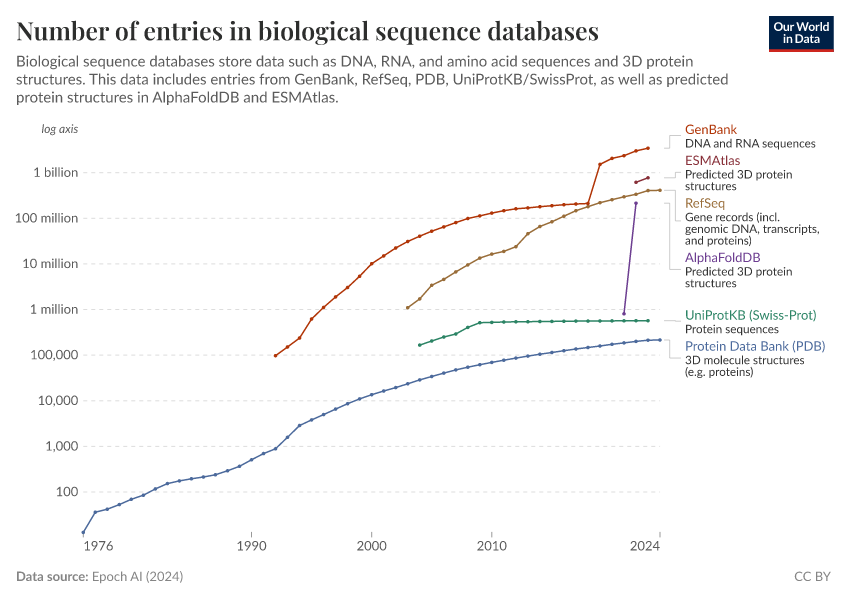
- GenBank is a database of RNA and DNA sequences maintained by the National Center for Biotechnology Information (NCBI). Researchers can submit nucleotide sequences, which are then reviewed and annotated by NCBI staff. Researchers are responsible for the scientific accuracy of their submissions.
- RefSeq is a curated collection of DNA, RNA, and protein sequences maintained by the NCBI. It provides reference sequences for major research organisms, including humans, model organisms, and pathogens. RefSeq records integrate information on genomic DNA, transcripts, and proteins to provide a complete picture of each gene in each organism.
- Protein Data Bank (PDB) is a database of 3D structural data of large biological molecules, such as proteins, nucleic acids, lipids, carbohydrates, and complex assemblies of these molecules. It only includes experimentally validated structures and is managed by the Research Collaboratory for Structural Bioinformatics (RCSB), which reviews each submission for quality and accuracy before adding it to the database.
- UniProtKB/Swiss-Prot is a database of protein sequence and functional information. This includes information on both the protein sequence and structure, as well as its effects and interactions in an organism. The Swiss-Prot section is manually curated and contains only experimentally verified information.
- AlphaFoldDB is a database of predicted 3D structures of proteins generated by the AlphaFold AI model, which uses deep learning to predict protein structures based on their amino acid sequences. While structures are generally not experimentally validated, when they are, predictions have been shown to be highly accurate.
- ESMAtlas is a database of predicted 3D structures of proteins generated by the ESM AI model, which predicts protein structures based on their amino acid sequences. While structures are generally not experimentally validated, when they are, predictions have been shown to be highly accurate.
- All databases listed here are freely accessible to the public and are widely used by researchers, educators, and students worldwide for various purposes, including scientific research, drug discovery, and education.
What you should know about this indicator
- Biological sequence data includes data such as DNA and RNA sequences, amino acid sequences of proteins, and three-dimensional structures of proteins and other molecules found in living organisms. These organisms can be bacteria, viruses, plants, animals and humans.
- Researchers use this data to better understand the biology of organisms, including the functions of genes and proteins and how they interact. This knowledge can then be applied to e.g. develop new drugs and treatments for illnesses or use biotechnology to improve agriculture and environmental science.
- This dataset provides an overview of the growth of key biological sequence databases over time.
- GenBank is a database of RNA and DNA sequences maintained by the National Center for Biotechnology Information (NCBI). Researchers can submit nucleotide sequences, which are then reviewed and annotated by NCBI staff. Researchers are responsible for the scientific accuracy of their submissions.
- RefSeq is a curated collection of DNA, RNA, and protein sequences maintained by the NCBI. It provides reference sequences for major research organisms, including humans, model organisms, and pathogens. RefSeq records integrate information on genomic DNA, transcripts, and proteins to provide a complete picture of each gene in each organism.
- Protein Data Bank (PDB) is a database of 3D structural data of large biological molecules, such as proteins, nucleic acids, lipids, carbohydrates, and complex assemblies of these molecules. It only includes experimentally validated structures and is managed by the Research Collaboratory for Structural Bioinformatics (RCSB), which reviews each submission for quality and accuracy before adding it to the database.
- UniProtKB/Swiss-Prot is a database of protein sequence and functional information. This includes information on both the protein sequence and structure, as well as its effects and interactions in an organism. The Swiss-Prot section is manually curated and contains only experimentally verified information.
- AlphaFoldDB is a database of predicted 3D structures of proteins generated by the AlphaFold AI model, which uses deep learning to predict protein structures based on their amino acid sequences. While structures are generally not experimentally validated, when they are, predictions have been shown to be highly accurate.
- ESMAtlas is a database of predicted 3D structures of proteins generated by the ESM AI model, which predicts protein structures based on their amino acid sequences. While structures are generally not experimentally validated, when they are, predictions have been shown to be highly accurate.
- All databases listed here are freely accessible to the public and are widely used by researchers, educators, and students worldwide for various purposes, including scientific research, drug discovery, and education.
Sources and processing
This data is based on the following sources
How we process data at Our World in Data
All data and visualizations on Our World in Data rely on data sourced from one or several original data providers. Preparing this original data involves several processing steps. Depending on the data, this can include standardizing country names and world region definitions, converting units, calculating derived indicators such as per capita measures, as well as adding or adapting metadata such as the name or the description given to an indicator.
At the link below you can find a detailed description of the structure of our data pipeline, including links to all the code used to prepare data across Our World in Data.
Notes on our processing step for this indicator
We use the data collected by Epoch AI on the growth of key biological sequence databases over time. We have added their extraction notes below for reference.
We show the maximum number of entries reported for each database in a given year.
Extraction notes from Epoch AI
- GenBank: Data extracted from release notes of GenBank.
- RefSeq: Data extracted from RefSeq release notes.
- UniProt: Data extracted from UniProt release notes and supplementary data from UniProt paper.
- PDB: Data extracted from RCSB PDB growth statistics webpage.
- AlphaFoldDB: Data extracted from AlphaFoldDB release notes and associated paper.
- ESMAtlas: Data extracted from ESMAtlas database information.
Reuse this work
- All data produced by third-party providers and made available by Our World in Data are subject to the license terms from the original providers. Our work would not be possible without the data providers we rely on, so we ask you to always cite them appropriately (see below). This is crucial to allow data providers to continue doing their work, enhancing, maintaining and updating valuable data.
- All data, visualizations, and code produced by Our World in Data are completely open access under the Creative Commons BY license. You have the permission to use, distribute, and reproduce these in any medium, provided the source and authors are credited.
Citations
How to cite this page
To cite this page overall, including any descriptions, FAQs or explanations of the data authored by Our World in Data, please use the following citation:
“Data Page: Number of entries in biological sequence databases”. Our World in Data (2026). Data adapted from Epoch AI. Retrieved from https://archive.ourworldindata.org/20251028-170323/grapher/number-of-entries-in-biological-sequence-databases.html [online resource] (archived on October 28, 2025).How to cite this data
In-line citationIf you have limited space (e.g. in data visualizations), you can use this abbreviated in-line citation:
Epoch AI (2024) – with major processing by Our World in DataFull citation
Epoch AI (2024) – with major processing by Our World in Data. “Number of entries in biological sequence databases” [dataset]. Epoch AI, “Trends in Biological Sequence Data” [original data]. Retrieved January 3, 2026 from https://archive.ourworldindata.org/20251028-170323/grapher/number-of-entries-in-biological-sequence-databases.html (archived on October 28, 2025).